k-Means Threading#
Requirements#
import pandas as pd
import numpy as np
from sklearn.preprocessing import OrdinalEncoder
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import resample
import time
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import MinMaxScaler
import threading
def standard_encoding(X) :
standard_encoding_ = OrdinalEncoder().fit_transform(X)
return standard_encoding_
k-Means class#
class fastkmeans :
def __init__(self, k, X, random_seed, n_iter) :
self.n_iter = n_iter
n = len(X)
if isinstance(X, pd.DataFrame):
X = X.to_numpy()
elif isinstance(X, np.ndarray):
pass
def initial_clusters() :
sample_index = resample(range(0, n), n_samples=n, replace=False, random_state=random_seed)
sample_index = np.array(sample_index)
size_Cr = round(n/k)
size_Cr_arr = np.repeat(size_Cr, k-1)
size_Ck = n - np.sum(size_Cr_arr)
size_Cr_list = list(size_Cr_arr)
size_Cr_list.append(size_Ck)
cluster_labels = np.array([])
for x in zip(range(0,k), size_Cr_list) :
cluster_labels = np.concatenate([cluster_labels, np.repeat(x[0], x[1])])
cluster_labels = np.array([int(x) for x in cluster_labels])
cluster_labels = cluster_labels[np.argsort(sample_index)]
return cluster_labels
def get_centroid(r, cluster_labels) :
obs_index = np.arange(n)
Cr_obs = obs_index[cluster_labels == r]
X_Cr = X[Cr_obs,:]
if len(X_Cr) > 0 :
centroid_Cr = X_Cr.mean(axis=0)
else :
centroid_Cr = X.mean(axis=0)
return centroid_Cr
def get_centroids(cluster_labels) :
centroids = [get_centroid(r, cluster_labels) for r in np.arange(k)]
return centroids
def intra_cluster_var(r, cluster_labels):
Cr_obs = np.where(cluster_labels == r)[0]
centroids = get_centroids(cluster_labels)
X_Cr = X[Cr_obs, :]
dist_Cr_obs = np.linalg.norm(X_Cr - centroids[r], axis=1)
var_Cr = np.sum(dist_Cr_obs**2)
return var_Cr
def WCSS(cluster_labels) :
intra_cluster_var_arr = np.array([intra_cluster_var(r, cluster_labels) for r in np.arange(k)])
WCSS_ = np.sum(intra_cluster_var_arr)
return WCSS_
def get_new_labels(centroids):
dist_obs_centroids = np.linalg.norm(X[:, np.newaxis, :] - centroids, axis=2)
cluster_labels = np.argmin(dist_obs_centroids, axis=1)
return cluster_labels
self.initial_clusters = initial_clusters
self.get_centroids = get_centroids
self.get_new_labels = get_new_labels
self.WCSS = WCSS
def fit(self, ) :
initial_clusters = self.initial_clusters
get_centroids = self.get_centroids
get_new_labels = self.get_new_labels
WCSS = self.WCSS
n_iter = self.n_iter
WCSS_dict = dict()
centroids_dict = dict()
labels_dict = dict()
b = n_iter - 1
for iter in range(0, b + 1) :
if iter == 0 :
labels_dict[iter] = initial_clusters()
centroids_dict[iter] = get_centroids(labels_dict[iter])
WCSS_dict[iter] = WCSS(labels_dict[iter])
else :
labels_dict[iter] = get_new_labels(centroids_dict[iter-1])
centroids_dict[iter] = get_centroids(labels_dict[iter])
WCSS_dict[iter] = WCSS(labels_dict[iter])
# Stop criteria
if iter >= 2 :
cond1 = np.isclose(WCSS_dict[iter], WCSS_dict[iter-1], atol=0.00005)
cond2 = np.isclose(WCSS_dict[iter], WCSS_dict[iter-2], atol=0.00005)
if cond1 and cond2 :
max_iter = iter + 1
break
else:
max_iter = n_iter
final_clusters_df = pd.DataFrame({'cluster_labels' : labels_dict[max_iter-1]})
self.WCSS_dict = WCSS_dict
self.centroids_dict = centroids_dict
self.labels_dict = labels_dict
self.max_iter = max_iter
self.final_clusters_df = final_clusters_df
Loading and processing the data#
computers_1000000 = pd.read_csv(r'C:\Users\fscielzo\Documents\DataScience-GitHub\Parallel Programming\Tech Lab 1\Data\computers_1000000.csv')
computers_1000000['cd'] = standard_encoding(computers_1000000[['cd']]) # no=0, yes=1
computers_1000000['laptop'] = standard_encoding(computers_1000000[['laptop']]) # no=0, yes=1
computers_1000000.drop(columns=['id'], inplace=True) # Remove 'id' column
Elbow computation with threading (parallelized)#
def compute_elbow(k, WCSS_Elbow_dict):
fastkmeans_init = fastkmeans(k=k, X=computers_1000000, random_seed=123, n_iter=100)
fastkmeans_init.fit()
WCSS_k = fastkmeans_init.WCSS_dict[fastkmeans_init.max_iter - 1]
WCSS_Elbow_dict[k] = WCSS_k
if __name__ == "__main__":
start_time = time.time()
k_values = range(2, 13)
WCSS_Elbow_dict = dict()
# Create threads and run compute_elbow for each k concurrently
threads_list = []
for k in k_values:
thread = threading.Thread(target=compute_elbow, args=(k, WCSS_Elbow_dict))
threads_list.append(thread)
thread.start()
# Wait for all threads to finish
for thread in threads_list:
thread.join()
end_time = time.time()
Time Elbow computation: 367.73306345939636
print('Time Elbow computation:', np.round((end_time - start_time)/60,2), 'mins')
Time Elbow computation: 6.13 mins
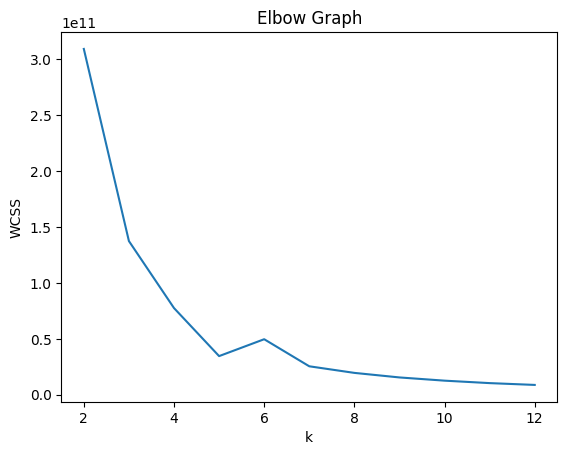
Elbow Graph#
k_values = range(2, 13)
WCSS_Elbow_values = list(WCSS_Elbow_dict.values())
sns.lineplot(x=k_values, y=WCSS_Elbow_values)
plt.xlabel("k")
plt.ylabel("WCSS")
plt.title("Elbow Graph")
plt.show()
Selection of optimal k#
# Calculate the first derivative
first_derivative = np.gradient(WCSS_Elbow_values)
# Calculate the second derivative
second_derivative = np.gradient(first_derivative)
# Plot the Elbow graph and second derivative
plt.figure(figsize=(12, 4))
plt.subplot(1, 3, 1)
plt.plot(k_values, WCSS_Elbow_values, marker='o')
plt.title("Elbow Graph")
plt.xlabel("Number of Clusters (k)")
plt.ylabel("WCSS")
plt.subplot(1, 3, 2)
plt.plot(k_values, second_derivative, marker='o', color='green')
plt.title("Second Derivative")
plt.xlabel("Number of Clusters (k)")
plt.ylabel("Second Gradient")
plt.tight_layout()
plt.show()
# Computing optimal k
k_optimal = k_values[np.argmax(second_derivative)]
print('Optimal k:' , k_optimal)
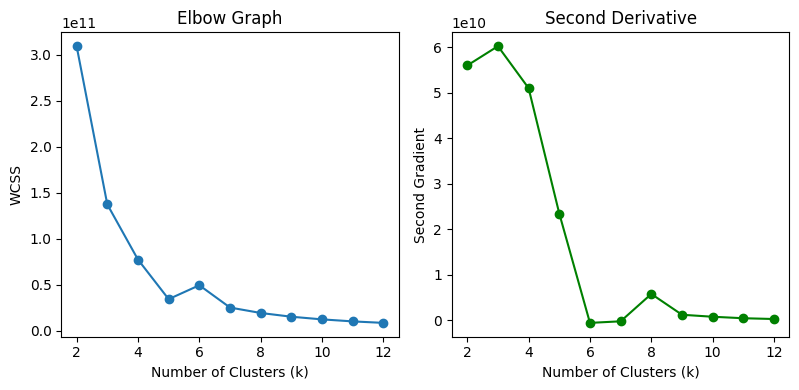
Optimal k: 3
Training k-means with optimal k#
start_time = time.time()
fastkmeans_init = fastkmeans(k=k_optimal, X=computers_1000000, random_seed=123, n_iter=100)
fastkmeans_init.fit()
end_time = time.time()
print('Training k-means with optimal k (for 1.000.000 rows):', end_time - start_time)
Training k-means with optimal k (for 1.000.000 rows): 4.2514472007751465
Computing the cluster with higher average price#
clusters_df = fastkmeans_init.final_clusters_df
avg_price_clusters = dict()
for r in range(0, k_optimal) :
index_Cr = clusters_df.loc[clusters_df['cluster_labels'] == r].index
avg_price_clusters[r] = computers_1000000.loc[index_Cr, 'price'].mean()
avg_price_clusters_list = list(avg_price_clusters.values())
cluster_higher_price = np.argmax(avg_price_clusters_list)
print('Cluster with higher average price:', cluster_higher_price)
print('Average price:', avg_price_clusters_list[cluster_higher_price])
Cluster with higher average price: 1
Average price: 3709.3009965920273
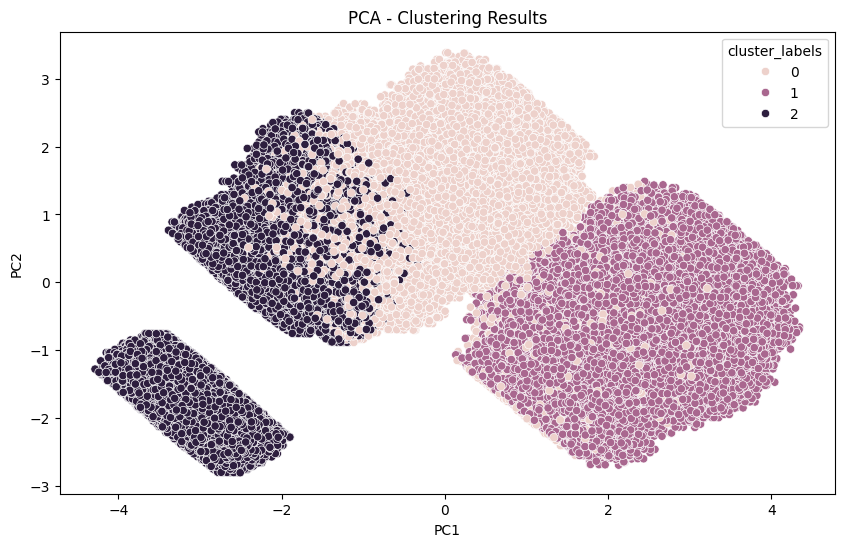
Plotting two first PCA dimensions#
X = computers_1000000
clusters_df = fastkmeans_init.final_clusters_df
scaler = StandardScaler()
X_std = scaler.fit_transform(X)
pca = PCA(n_components=2)
X_pca = pca.fit_transform(X_std)
X_pca_df = pd.DataFrame(X_pca, columns=['PC1', 'PC2'])
X_pca_df['cluster_labels'] = clusters_df['cluster_labels']
plt.figure(figsize=(10, 6))
sns.scatterplot(x='PC1', y='PC2', hue='cluster_labels', data=X_pca_df)
plt.xlabel('PC1')
plt.ylabel('PC2')
plt.title('PCA - Clustering Results')
plt.show()
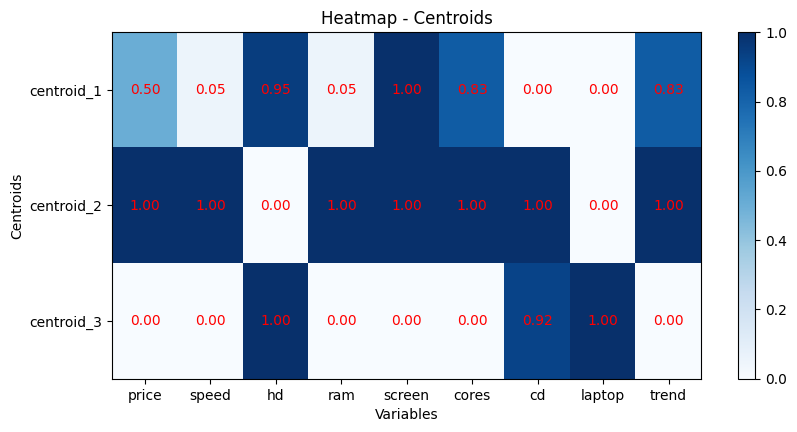
Plotting a HeatMap#
scaler = MinMaxScaler()
centroids_arr = np.array(fastkmeans_init.centroids_dict[fastkmeans_init.max_iter-1])
scale_centroids_arr = scaler.fit_transform(centroids_arr)
scale_centroids_df = pd.DataFrame(scale_centroids_arr)
scale_centroids_df.index = ['centroid_1', 'centroid_2', 'centroid_3']
scale_centroids_df.columns = computers_1000000.columns
plt.figure(figsize=(9.5, 4.5))
cax = plt.imshow(scale_centroids_df, cmap='Blues', aspect='auto')
for j in range(0,scale_centroids_df.shape[1]):
plt.text(j, 0, f'{scale_centroids_df.iloc[0, j]:.2f}', ha='center', va='center', color='red')
for i in range(1,scale_centroids_df.shape[0]):
for j in range(0,scale_centroids_df.shape[1]):
plt.text(j, i, f'{scale_centroids_df.iloc[i, j]:.2f}', ha='center', va='center', color='red')
plt.colorbar(cax)
plt.title('Heatmap - Centroids')
plt.ylabel('Centroids')
plt.xlabel('Variables')
plt.xticks(ticks=range(len(scale_centroids_df.columns)), labels=scale_centroids_df.columns)
plt.yticks(ticks=range(len(scale_centroids_df.index)), labels=scale_centroids_df.index)
plt.show()